Chinese and Swiss Scientists Decipher the Spatial Code of Root Microbial Colonization

Cover Story
Selective recruitment of soil bacteria by plants is critical for the assembly of a highly specialized rhizosphere microbiome. Root bacterial communities influence root development and plant health and confer resilience against a range of biotic and abiotic stresses. Plants shape their microbial communities through the release of root exudates-a complex mixture of organic compounds. Despite an understanding that exudates are central to bacterial colonization, little is known about how, when, and where they are released, especially at the spatial scale relevant for bacteria.
On October 3, 2025, Professor Feng Zhou’s team in collaboration with Professor Niko Geldner's group at the University of Lausanne, Switzerland, published a landmark cover paper in Science entitled "Localized glutamine leakage drives the spatial structure of root microbial colonization", which unveiled how plant roots guide microbes to specific sites, mapping a unprecedented colonization patterns of root-associated microbes.
This research first found that bacterial colonization patterns coincided with absent or impaired endodermal Casparian strips suggesting that transient endodermal leakages of organic compounds might drive bacterial attraction and colonization of roots. Mass spectrometry analysis of root exudates from Casparian strip–defective mutants revealed that vasculature-derived glutamine is a major bacterial chemoattractant. Using a combination of genetic manipulation and reporters in both plant and the commensal model bacterium Pseudomonas protegens CHA0, this study demonstrated that glutamine leakage is a major cue for bacteria to find and colonize precise sites on the root, especially at the lateral root emergence sites.
Moreover, this study showed that bacteria metabolically adapt to this glutamine-rich niche and use glutamine as a carbon source, allowing their increased proliferation. It has also been shown that alterations of bacterial abundance within complex synthetic communities in response to endodermal leakage can partially be explained by the ability of bacteria to use glutamine as a carbon source. Associated chronic immune stimulation suggested that endodermal nutrient restriction is crucial for regulating microbial colonization and assembly, limiting excessive proliferation that could compromise plant health.
This study provides evidence for transient glutamine leakages from the vasculature being an important driver of bacterial colonization. This identifies transient metabolite leakage as a third path of root exudate formation, complementing the previously proposed mechanisms of controlled secretion or transporter-mediated export and the uncontrolled release of cellular contents during root cap shedding. By integrating plant developmental biology with highly resolved microbial colonization studies, this study have elucidated the crucial role for localized exudate leakages in shaping microbial assembly along the complex and rapidly developing root surfaces. These findings illustrate dynamic root-microbe interactions in which microbial communities are “seeded” by providing short-lived and spatially restricted metabolic niches. Understanding these dynamics in crops could ultimately open new strategies for microbiome-informed breeding or management practices to enhance stress resilience and plant health.
Professor Feng Zhou from the CAS Center for Excellence in Molecular Plant Sciences (CEMPS) and Professor Niko Geldner from the Department of Plant Molecular Biology at the University of Lausanne (UNIL) acted as co-corresponding authors, while doctoral student Yuanjie Tang from the CEMPS and postdoctoral researcher Huei-Hsuan Tsai from the UNIL are co-first authors of the paper. This research was supported by funds from the National Key R&D Program of China, the Strategic Priority Research Program of the Chinese Academy of Sciences, a European Research Council grant.
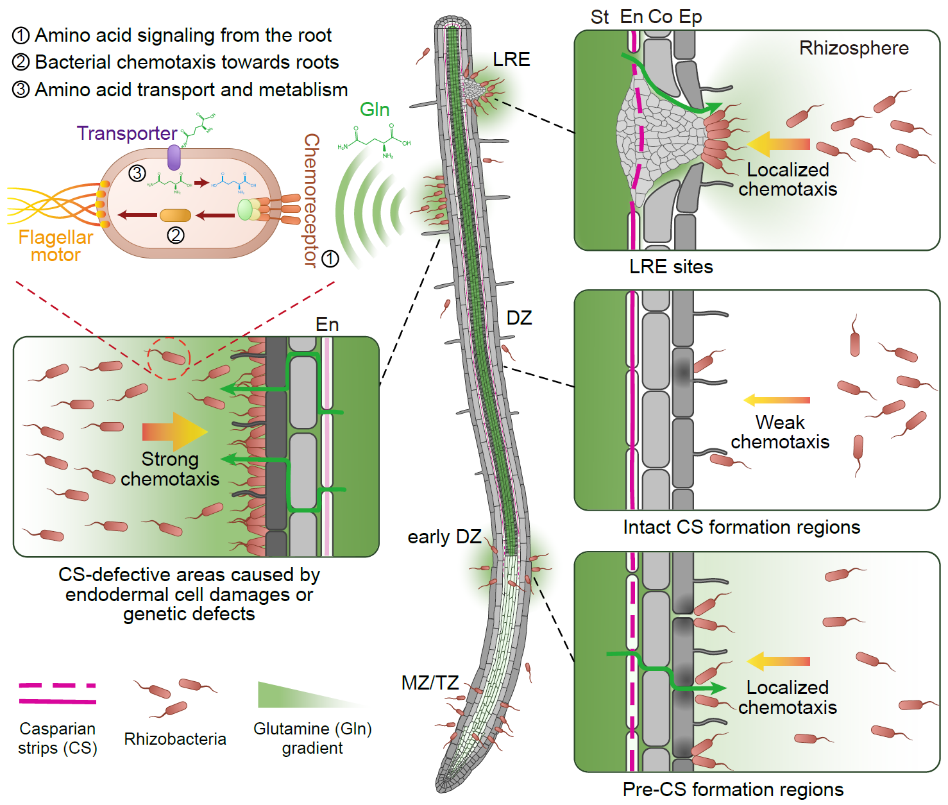
Localized glutamine leakage controls spatial colonization patterns of rhizobacteria
Link:https://www.science.org/doi/10.1126/science.adu4235
Contact: http://fzhou@cemps.ac.cn